About

Hello! I am data driven scientist currently working as a postdoctoral
research scientist at Columbia University Irving Medical Center under Dr. Claudia Doege and Prof. Rudy Leibel.
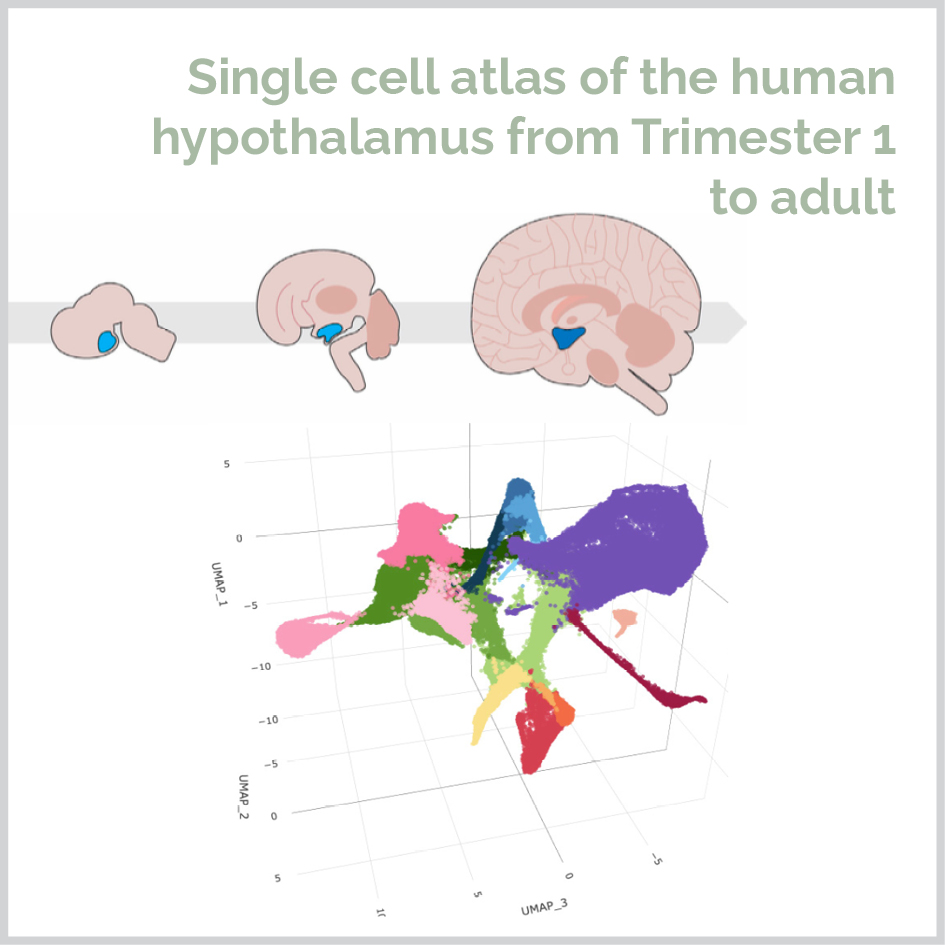
My Postdoc research focuses on using next generation sequencing data from the hypothalamus to understand the genetic mutations underpinning feeding circuit abnormalities leading to obesity. I work with large datasets including single-cell RNAseq, multiome, bulk RNAseq, and spatial transcriptomics, to conduct in depth clustering, characterize cell populations, generate pseduotime trajectories, identify transcription factor signatures and prioritize genes for CRISPRi screening. I developed the first atlas of human hypothalamic development spanning Trimester 1 through adulthood. I generated a new R package – Glowworm – as a tool for researchers to use single cell transcriptomics data to prioritize disease genes based on the cell types within their data.
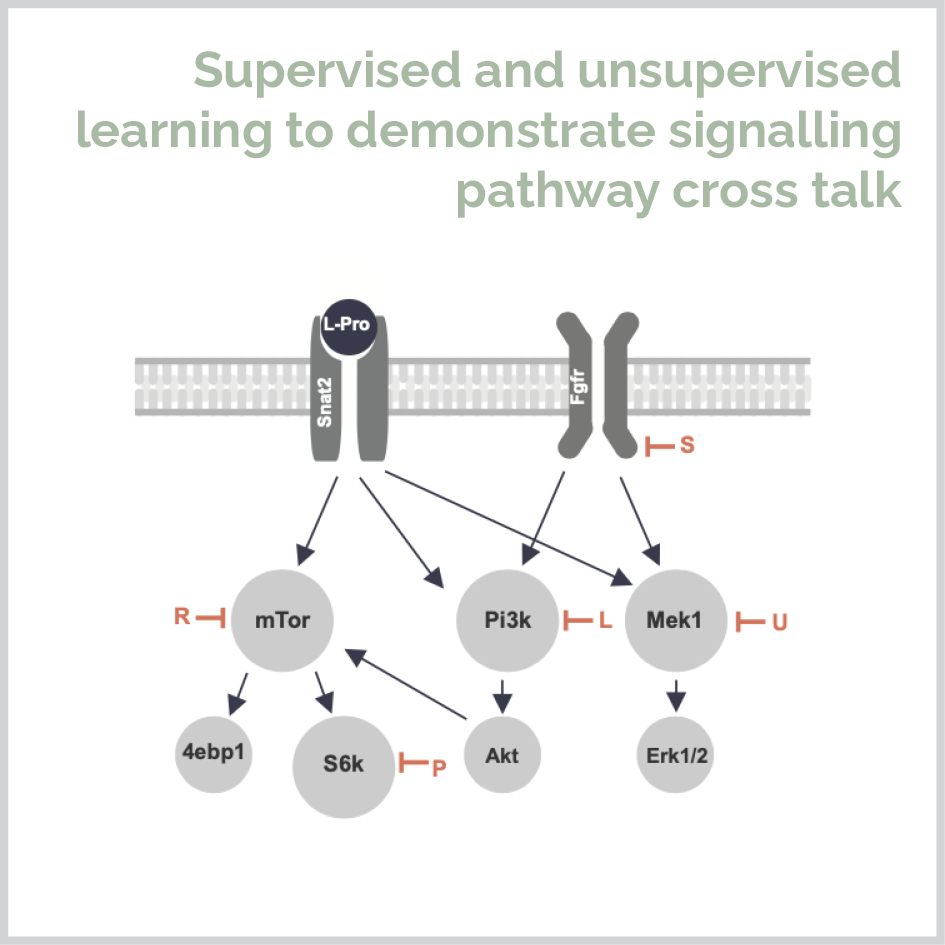
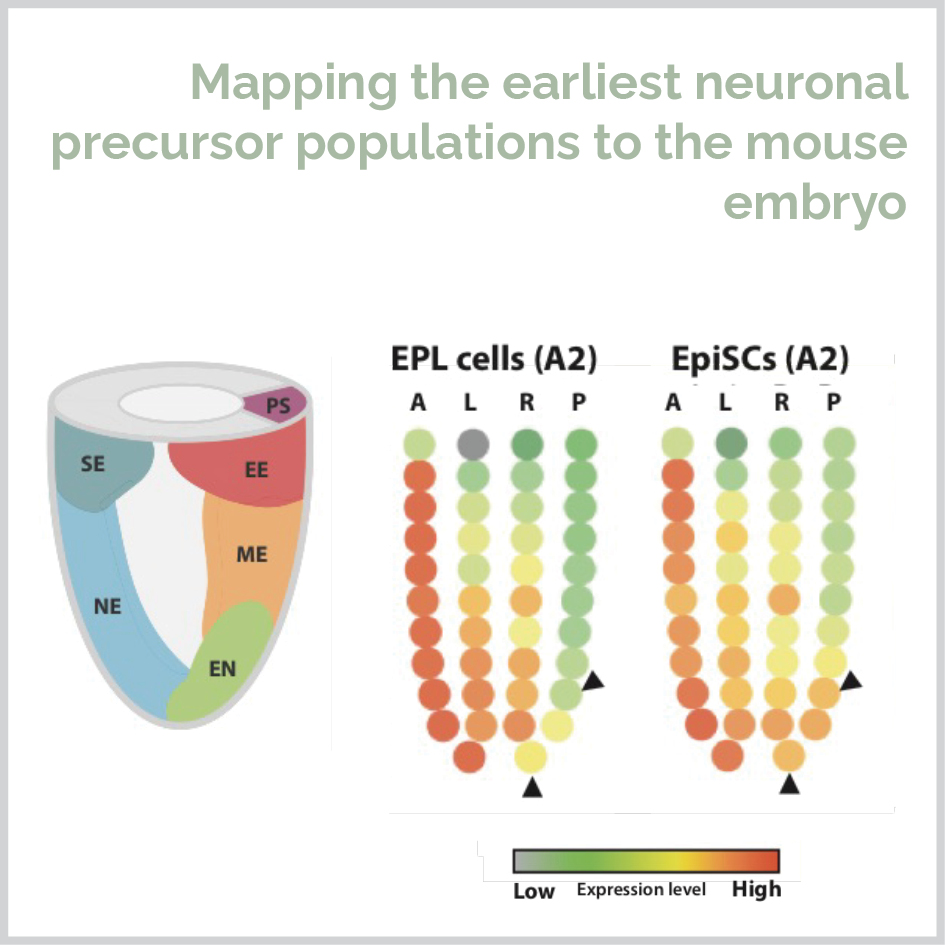
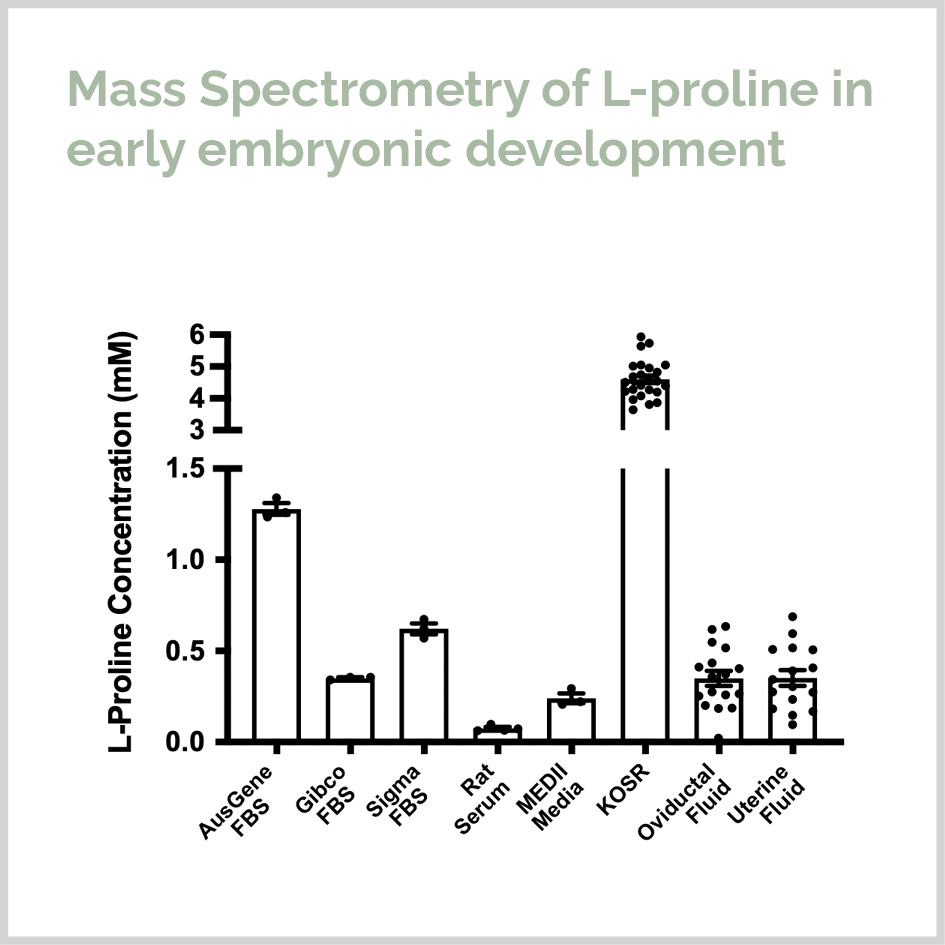
I completed my PhD at the University of Sydney in 2019 under the supervision of Dr. Michael Morris and Margot Day. During my PhD I combined wet lab science, bioinformatics and statistical modelling to study the mechanism of L-proline in stem cell pluripotency and differentiation to neurons. I used unsupervised, supervised and machine learning approaches to show that the amino acid L-proline activates five signaling pathways to initiate drive pluripotent cells to form a genetically and spatially distinct cell population corresponding to the in vivo neurectoderm.
My Postdoc research focuses on using next generation sequencing data from the hypothalamus to understand the genetic mutations underpinning feeding circuit abnormalities leading to obesity. I work with large datasets including single-cell RNAseq, multiome, bulk RNAseq, and spatial transcriptomics, to conduct in depth clustering, characterize cell populations, generate pseduotime trajectories, identify transcription factor signatures and prioritize genes for CRISPRi screening. I developed the first atlas of human hypothalamic development spanning Trimester 1 through adulthood. I generated a new R package – Glowworm – as a tool for researchers to use single cell transcriptomics data to prioritize disease genes based on the cell types within their data.
I completed my PhD at the University of Sydney in 2019 under the supervision of Dr. Michael Morris and Margot Day. During my PhD I combined wet lab science, bioinformatics and statistical modelling to study the mechanism of L-proline in stem cell pluripotency and differentiation to neurons. I used unsupervised, supervised and machine learning approaches to show that the amino acid L-proline activates five signaling pathways to initiate drive pluripotent cells to form a genetically and spatially distinct cell population corresponding to the in vivo neurectoderm.
Technical Skills
- R Programming: tidyverse/dplyr, Seurat, Signac, ggplot2, Monocle3, Shiny, igraph, Bioconductor, car, MASS
- Python Programming: numpy, pandas, matplotlib, CELLECT, scikit-learn
- Data Visualization: Adobe suite, ggplot2 and Tableau.
- Statistical: Unsupervised and supervised learning; Statistical methodology including inferential statistics and hypothesis testing
- Other Computational: Cloud computing using Linux OS on Amazon Web Server (AWS); SQL management; Git versioning; Command-line interface (CLI) including NGS alignment (Kallisto, TopHat, STAR, Bowtie2); Working with VCF, BAM/BED and FASTQ files; Working with genetic databases including dbSNP, RefSeq, Ensembl and NCBI
- Wet Lab: Cell culture; qRT-PCR; RNAseq; Flow cytometry; Microscopy; Mass spectrometry; Western blotting; ELISA.
Publications
For a full list of my publications, visit my Google Scholar page
Contact
Email:
hannahjglover@gmail.com